CORONA (COvid19 Registry of Off-label & New Agents) Project
Director/Lead Investigator: David Fajgenbaum, MD, MBA, MSc
About the Project
The CDCN launched the CORONA Project in March 2020 to identify and track all treatments reported to be used for COVID-19 in an open-source data repository. The CORONA team has reviewed 29,353 papers and extracted 2,399 papers on 590 treatments administered to 437,936 patients with COVID-19, and in continuing to help researchers to prioritize treatments for clinical trials and inform patient care.
All data is made available via our Tableau-based CORONA Viewer that has recorded over 23,000 views to date and counts staff at Google Health, HHS, FDA, and NIH among regular users. You can visit the CORONA database viewer built and managed by Matt Chadsey, owner of Nonlinear Ventures below.
We have also written some high level articles about the project and its findings in our CORONA INSIGHTS blog.
Goals
The overarching goal of the CORONA project is to advance effective treatments for COVID-19 by highlighting the most promising treatments to pursue, informing optimal clinical trial study design (sample size, target subpopulations), and determining if a drug should move forward to widespread clinical use. While there have been several notable failures in drug repurposing for COVID-19, a handful of drugs, including dexamethasone, heparin and baricitinib, have likely helped save thousands of lives and more treatments are urgently needed for newly diagnosed and soon to- be-diagnosed COVID-19 patients while vaccinations are underway and potential SARS-CoV-2 variants emerge. We’ve identified ~10 additional promising medications that, in collaboration with researchers at the FDA, NIH and elsewhere, we’re hoping to move into randomized control trials.
To further pursue our goal by the end of 2021, we will be expanding our work to also integrate pre-clinical and randomized controlled trial data. We believe that this expanded focus will allow us to more quickly advance promising treatments to clinical care that are supported by broader research findings.
Key Milestones
- In May 2020, we completed a systematic review of treatments given to the first 9,152 COVID-19 patients reported in 2500+ papers that was published in Infectious Diseases and Therapy: Treatments Administered to the First 9152 Reported Cases of COVID-19: A Systematic Review – PennMedicine Press Release
- In spring 2020, dexamethasone was identified as a promising treatment approach from data in CORONA months before it was proven effective through a large clinical trial. Inhaled interferon was similarly identified as a promising agent before a trial demonstrated its effectiveness.
- In spring 2020, Dr. Fajgenbaum highlighted the CORONA Project’s progress and lessons from Chasing My Cure on the following outlets: NPR’s Fresh Air, 6ABC, KTLA, FOX5, ABC7, and WRAL.
- In summer 2020, the CORONA Project was profiled by the following outlets: Boston Globe, ABC’s 20/20, and CNN.
- In July 2020, Dr. Fajgenbaum presented about the CORONA Project during the University of Pennsylvania’s COVID-19 Symposium.
- In fall 2020, JAK1/2 inhibition was identified as a promising approach for severe COVID-19 through research in the CSTL (paper under review). The JAK1/2 inhibitor baricitinib was subsequently found to be effective in a large clinical trial.
- In fall 2020, we launched CORONA Insights, a page where we will periodically share posts summarizing key findings uncovered in the CORONA database.
- In December 2020, CORONA Director, Dr. David Fajgenbaum, co-authored an article in the New England Journal of Medicine on Cytokine Storm that involves the immune system attacking vital organs. This can occur in Castleman disease and COVID-19.
- In January 2021, CORONA partnered with the Parker Institute for Cancer Immunotherapy to expand its scope. See the press release here.
FAQ
What data sources are used?
- Data regarding clinical trial registrations come from international trial registration websites, such as clinicaltrials.gov. Information from these websites are aggregated by COVID-NMA and the aggregated data are exported for use in the CORONA Project.
- COVID-NMA: Thu Van Nguyen, Gabriel Ferrand, Sarah Cohen-Boulakia, Ruben Martinez, Philipp Kapp, Emmanuel Coquery, … for the COVID-NMA consortium. (2020). RCT studies on preventive measures and treatments for COVID-19 [Data set]. Zenodo. http://doi.org/10.5281/zenodo.4266528
- Information about treatment guidelines, such as those issues by the National Institutes of Health, are found on the website of the institutions providing guidelines.
- Data from published papers comes directly from the publications, as extracted by the CORONA Project’s data coordinators. Data are extracted as provided by the studies’ authors. However, errors may occur. Please report any errors or concerns to tsikora@pennmedicine.upenn.edu.
What is the Research Prioritization grade?
The Research Prioritization (RP) grade is based on evidence from published randomized controlled trials. An evidence synthesis is performed for each drug or combination of drugs. If drugs are used in both the inpatient and outpatient settings, separate evidence syntheses are performed for each clinical setting. Each drug-setting receives an overall RP grade consisting of 1) a treatment effect assessment (likely beneficial, benefit unknown, not likely beneficial) and 2) certainty of evidence assessment (high certainty, moderate/low certainty). These grades are presented as letter grades for simplified comparison. The purpose of the RP grade is to identify promising drugs that require further evaluation.
What does the Research Prioritization grade indicate?
The purpose of the Research Prioritization grade is to identify promising drugs that require further evaluation. “A” treatments are likely beneficial with high certainty. They are generally well-studied and have a known likelihood of benefit. These treatments rarely need further research. “B” treatments are likely beneficial with moderate to low certainty. They have indicators of benefit, but this determination may be based on insufficient data. These treatments are well-positioned for further research to better characterize efficacy and should generally be prioritized for additional clinical trials. “B/C” treatments may have a trend towards benefit, but the data synthesis indicates that the benefit is not yet clear. “C” treatments are not likely beneficial with moderate to low certainty. They do not appear beneficial based on the available data, but there may be insufficient data to make a robust determination. These treatments are generally not prioritized for further research. “D” treatments are not likely beneficial with high certainty, based on a substantial amount of previous research. In general, “D” treatments do not require further research.
| Letter Grade | Definition |
| A | Likely beneficial with high certainty |
| B | Likely beneficial with moderate to low certainty |
| B/C | Grade uncertain |
| C | Not likely beneficial with moderate to low certainty |
| D | Not likely beneficial with high certainty |
What is the Treatment Efficacy grade?
The Treatment Efficacy (TE) grade is based on evidence from high-quality randomized controlled trials. By limiting data to well-run, peer-reviewed studies, this grade is intended to provide an assessment of the available evidence for use in a clinical setting. In contrast, the Research Prioritization grade includes pre-prints, small studies, etc. that may provide valuable signals about a treatment’s efficacy but are unlikely to be robust enough to guide treatment recommendations with high certainty. In addition to this algorithmic grade, information on international treatment guidelines, such as from the NIH and IDSA, are provided alongside for further context. Note that some treatments are recommended for particular subgroups, such as those on mechanical ventilation. If treatment guidelines recommend that a drug is best used in a particular setting, this will be noted in the CORONA viewer along with the treatment efficacy letter grade. The purpose of the TE grade is to serve as an indicator of whether sufficient data exist in support of using a treatment in clinical practice at this time for COVID-19.
How are the Research Prioritization and Treatment Efficacy grades formulated?
The grades are both based on two components, a likelihood of benefit assessment and a certainty of evidence assessment, as described below.
Likelihood of benefit assessment
The treatment effect portion of the overall grade is currently calculated using two methods:
- Fisher’s method p-value: Combining P-values of the primary endpoints of each study for a given treatment. (as described in https://training.cochrane.org/handbook/current/chapter-10) We analyze p-values across all studies using Fisher’s method, where Pi is the one-sided p-value for study i.
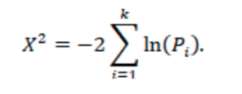 The test statistic follows a chi-squared distribution, which can be used to calculate a p-value for the null hypothesis that there is no evidence of an effect in at least one study. A p-value of <0.05 indicates that there is evidence of an effect in at least one study. Note that if a study provides a one-sided p-value, it is assumed that the hypothesis is in favor of the treatment under investigation.
The test statistic follows a chi-squared distribution, which can be used to calculate a p-value for the null hypothesis that there is no evidence of an effect in at least one study. A p-value of <0.05 indicates that there is evidence of an effect in at least one study. Note that if a study provides a one-sided p-value, it is assumed that the hypothesis is in favor of the treatment under investigation.
- Proportion of studies with beneficial effect direction: Summarizing the effect direction of the primary endpoints of each study. For example, if mortality is lower in the treatment group, the treatment is noted as having a “beneficial” effect for that given endpoint. (“Beneficial” is used only to indicate the direction of effect, not it’s magnitude or significance)
We are considering including other methods of synthesizing treatment effect, including a true meta-analysis or an analysis focusing on summarizing a select number of effect estimates. However, both methods would be best used if comparing identical endpoints across multiple studies – e.g. mortality. We are extracting multiple data points from all publications, so it is possible to explore the methods in the medium-term.
Certainty of evidence assessment
The certainty of evidence for a given treatment is currently determined by looking at three metrics:
- Precision: Sample size across all peer-reviewed trials listed in PubMed included in the evidence synthesis (≥500 total patients treated is required for a high certainty of evidence). When calculating total sample size, we consider only articles that were peer-reviewed (to minimize the impact of erroneous results from pre-prints) and listed in PubMed (to minimize the impact of results from unreputable journals). However, pre-prints and those not listed in Pubmed are still used for drug effect estimate analysis
- Directness of evidence: Number of trials included in the evidence synthesis compared to placebo or standard of care. ≥50% of trials included in the evidence synthesis must be compared to placebo or standard of care for a high certainty of evidence.
- Publication quality: Number of trials included in the evidence synthesis published in a journal that is indexed in PubMed. ≥75% of trials included in the evidence synthesis must be published in a journal that is indexed in PubMed for a high certainty of evidence.
The certainty of evidence for a given treatment is determined by incorporating each of the factors above. A “high” certainty of evidence assessment is for treatments that meet the threshold for all the factors above. A “moderate” certainty of evidence is reserved for treatments that meet some but not all the thresholds, and a “low” certainty of evidence is assigned to treatments that meet none.
What does a trial ID starting with XXX indicate?
Trials that have a trial ID beginning with XXX were not known to have a trial registration number. This may be because the published paper did not indicate a trial registration number and data coordinators could not link it to a known trial. It may also occur if a trial was run without being registered. Errors may occur during the data extraction process. Please report any errors or concerns to tsikora@pennmedicine.upenn.edu.
How can the data be used?
Data may be used freely and no permission is required. However, we do request attribution when using the manually extracted data (including study endpoint values, p-values, effect estimates, etc.).
Who should I contact with questions or concerns?
The data in this project are extracted manually from published papers by data coordinators. Errors may occur. Please report any errors or concerns to tsikora@pennmedicine.upenn.edu.



